...
For mouse, you have the option to choose a TSS file, containing either a broad or narrow definition of the genes’ TSS (Transcription Start Site) regions (based on <Author et al>, Nature. 2016 Jun 30;534(7609):652-7 - The landscape of accessible chromatin in mammalian preimplantation embryos).
...
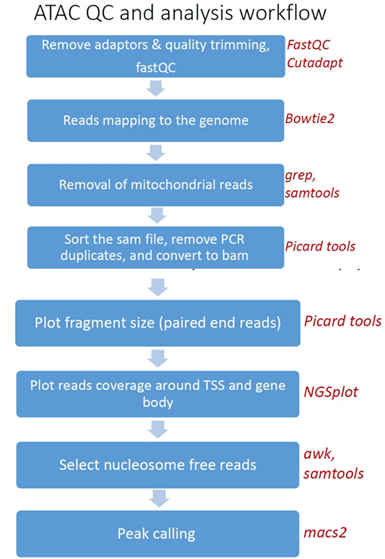
- Reads trimming: Reads are quality trimmed using cutadapt. In this process primers corresponding to the TruSeq protocol are removed (output is in folder 1).
- Quality control: Reads quality control is evaluated using FastQC (in output folder 2), and a report file, containing quality reports for all of the samples, is generated using multiQC (in output folder 3).
- Mapping to genome: The quality trimmed paired-end reads are mapped to Mouse/Human genomes using Bowtie2 (output is in folder 4).
- Alignment filtering: Following the alignment, mitochondrial genes are removed from the analysis (using the grep command). Duplicated reads are removed using picard-tools. The remaining unique reads are indexed and sorted using samtools index and samtools sort. Statistics on the alignment is generated using flagstat (output is in folder 5).
- Select nucleosome-free fragments: fragments of length <120bp are selected using the awk command (alignments are in folder 6), and insert size distributions are plotted (output is in folder 68).
- Visualization in graphs: The analyzed reads reads coverage on gene body and around the TSS are graphically visualized using ngsplot (output is in folder 7).
- Read counts on TSS: for mm10 genome we count the number of reads on genes’ TSS (Transcription Start Site) regions based on, Nature. 2016 Jun 30;534(7609):652-7 ).
- Peak calling: Peaks Broad peaks are called using MACS2 (output is in folder 10).
Output folders:
1_cutadapt
...
4_mapping
5_process_alignment
6_nucleosome_free
7_ngs_plot7
8_nucleosomepicard_freeplot
89_tss_count
910_call_peak
1011_reports
Log files (one directory above the output directory):
...