16.2.22 UTAP: Ribo-Seq Pipeline
Pipeline to run Ribo-Seq analysis from sequences to UTR and CDS counts and MACS peaks to determine ribosome enrichment regions.
Developed by Dena Leshkowitz and programmed for the UTAP pipeline by Michal Twik and Jordana Lindner. Details regarding the pipeline are available here.
Pipeline website: http://utap.wexac.weizmann.ac.il
Setting up a new analysis
The transcriptome pipelines run on the Wexac cluster. In order to run a new transcriptome analysis, your fastq files must be in your Collaboration folder on Wexac, in the correct structure (UTAP requirements & description).
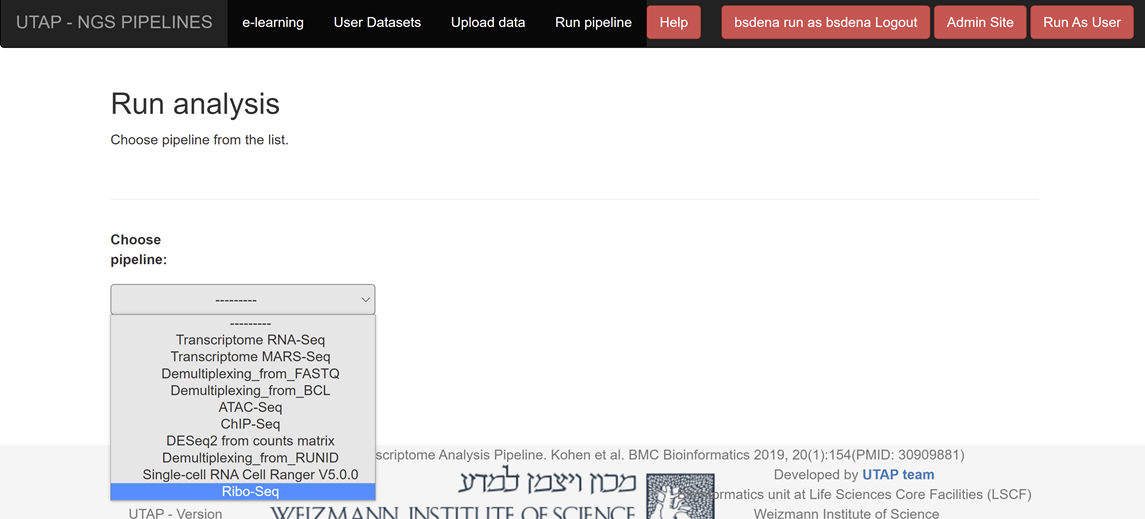
- Then, login to utap.wexac.weizmann.ac.il via Firefox or Chrome (the pipeline is NOT compatible with Internet Explorer) using your Weizmann userID and password, and click on Run pipeline:
Click on "Ribo-Seq" in the Choose pipeline box
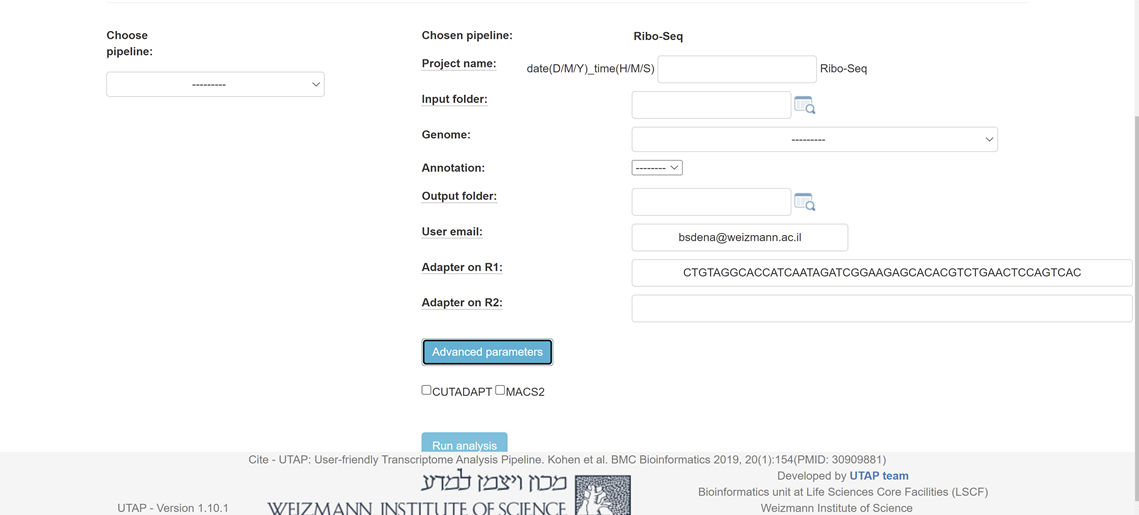
Please fill all the relevant information in the "choice box" .
LINK:
Transcriptome pipeline for Weizmann Institute users: http://utap.wexac.weizmann.ac.il
Acknowledgments
Citation:
Kohen et al. BMC Bioinformatics (2019) 20:154 https://doi.org/10.1186/s12859-019-2728-2 (PMID: 30909881)
Bioinformatics support staff for UTAP:
- UTAP development and maintenance team: utap@weizmann.ac.il
- Dena Leshkowitz
- Ester Feldmesser
- Gil Stelzer
- Bareket Dassa
- Noa Wigoda