NGS Support
Illumina sequencer run setup services (ngs-pipeline) and UTAP
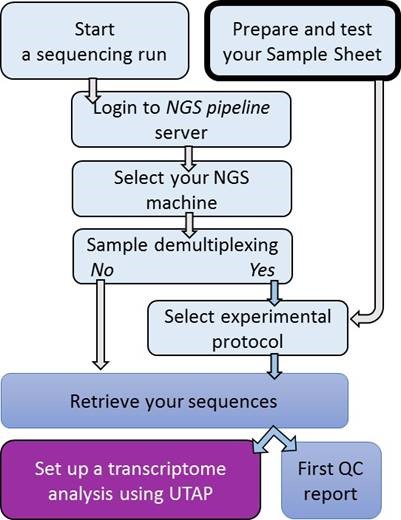
The bioinformatics unit provides several services for processing NGS data, divided into 2 main steps as follows:
NGS setup and services (ngs-pipeline) that assist with retrieving raw fastq or demultiplexed files, and provide quality control measures for each sample that was sequenced including:
Sequencing setup
Additional services for sequences
Our User-friendly Transcriptome Analysis Pipeline (UTAP) - Click to go to our UTAP site
- Analysis setup after sequencing completes
Analysis pipeline steps and reports
List of links
Sequencing pipeline:
https://ngs-pipeline.weizmann.ac.il/
Help page:
https://ngs-pipeline.weizmann.ac.il/ngsb/howto
QC:
http://stefan.weizmann.ac.il/fqc/{type RUN ID here}, for example:
http://stefan.weizmann.ac.il/raw/180509_NB551168_0120_AHTKN2BGX5/
List of runs, with delete functionality:
https://ngs-pipeline.weizmann.ac.il/ngsb/storage
Transcriptome pipeline:
http://ngspipe.wexac.weizmann.ac.il:7000
Demo of the UTAP interface:
UTAP - User-friendly Transcriptome Analysis Pipeline (for external users)
Acknowledgments
Citation:
Kohen et al. BMC Bioinformatics (2019) 20:154 https://doi.org/10.1186/s12859-019-2728-2 (PMID: 30909881)
Bioinformatics support staff for UTAP:
- UTAP team (UTAP maintenance) utap@weizmann.ac.il
- Dena Leshkowitz
- Ester Feldmesser
- Gil Stelzer
- Bareket Dassa
- Noa Wigoda